Over the last two articles of this series, we have discussed different Big Data file formats and their overall characteristics. We have also performed writing & reading examples using different Python modules & methods.
In this section, we will focus on comparing the performance of the formats reviewed. We will evaluate writing times, reading times, and file sizes.
We will then conclude this series by reviewing specific use cases for each one of the formats, as well as discussing some recommendations.
This section will be longer than the previous ones and involve more code. We’ll be using Python scripts which can be found in the Blog Article Repo.
Importing the required modules
For this section, we will be using the following Python libraries and modules:
Code
# File manipulation modules
import pandas as pd
from fastavro import reader, writer, parse_schema
import pickle
import openpyxl
# Performance measurement modules
import time
# System utility modules
import os
import shutil
from pathlib import Path
# Plotting modules
import matplotlib
import matplotlib.pyplot as plt
import seaborn as snsDefining plot parameters
Since we’ll be plotting our experimental results, we will need to define our plot parameters beforehand:
Code
# Before anything else, delete the Matplotlib
# font cache directory if it exists, to ensure
# custom font proper loading
try:
shutil.rmtree(matplotlib.get_cachedir())
except FileNotFoundError:
pass
# Define main color as hex
color_main = '#1a1a1a'
# Define title & label padding
text_padding = 18
# Define font sizes
title_font_size = 17
label_font_size = 14
# Define rc params
plt.rcParams['figure.figsize'] = [14.0, 7.0]
plt.rcParams['figure.dpi'] = 300
plt.rcParams['grid.color'] = 'k'
plt.rcParams['grid.linestyle'] = ':'
plt.rcParams['grid.linewidth'] = 0.5
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.sans-serif'] = ['Lora']Preparing the data set
To reduce noise in our performance measurements, we will employ a slightly larger file for this section. We will be using the Spotify Charts data set (2.48 GB) published by DHRUVIL DAVE, which you can download from Kaggle.
We can start by reading the data set as a pandas.DataFrame object:
Code
# Load csv file as pandas.DataFrame object
df = pd.read_csv('datasets/charts.csv')This process should take a couple of minutes, depending on the specifications of each machine.
Upon concluding, we should end up with a pandas.DataFrame object df with the following shape:
Code
# Get the shape of the object
df.shapeOutput
(26173514, 9)Meaning 26,173,514 rows by 9 columns.
Next, we will need to do some preprocessing before beginning with the tests:
Code
# Singe we have nan values, we will remove them
df = df.dropna()
# Check the current data types and see if casting is required
df.dtypes
# Cast to required data types
# Date is currently a string. We will cast it to Pandas DateTime in integer format.
# since Avro does not support original DateTime
df['date'] = pd.to_datetime(df['date'])
df['date'] = df['date'].apply(lambda x: x.value)
# Streams will be casted to integer type
df['streams'] = df['streams'].astype('int')
# Finally, reset index since .feather does not support
# serializing a non-default index
df = df.reset_index(drop = True)We converted our date field to an int datetime64 data type because the fastavro library does not currently support str datetime64 objects.
Since we will be defining a schema for this file format, a conventional datetime64 data type would raise a TypeError.
Lastly, we will create two new directories:
- The first one for storing all of our written files.
- The second one for storing all of our performance test results.
Code
mkdir performance_tests
mkdir performance_resultsNow, we’re ready to start designing our experiment.
Experiment design
For both writing & reading performance tests, we will be using a collection of measurements per file format. This is always a good practice in experimental design and will help us calculate a complete set of descriptive statistics.
Our experiment for the writing process will consist of the following steps:
- Define the variables to measure.
- Set the number of trials as a control variable.
- Begin with the first trial, measuring variables of interest.
- Store measurements.
- Repeat for the other file formats.
- Consolidate results and perform a statistical description.
- Delete all written files except one to read in the next experiment.
Our experiment for the reading process will consist of the following steps:
- Define the variables to measure.
- Set the number of trials as a control variable.
- Begin with the first trial, measuring variables of interest.
- Store measurements.
- Clear memory
- Repeat for the other file formats.
- Consolidate results and perform a statistical description.
Performance tests
1. Parameter definition
We will start by defining our experiment parameters:
Code
# Define number of trials n
n = 20
# Define performance tests output path
path = 'performance_tests/'2. Writing performance tests
We will start by defining our writingPerformance() function, which will accept the following parameters :
n:int- Number of trials.
path:str- Path for writing results.
df:pandas.DataFrame- DataFrame Object containing the data set.
Upon calling, it will return the following:
measured_vars_w:dict- Execution time for each file format with
nnumber of trials.
- Execution time for each file format with
Once we have the expected inputs and outputs, the idea is to perform the following:
- Declare a
pandas.core.series.Seriesfor each file format, where we will store writing times. This will result in 8 objects in total. - Declare an empty dictionary
measured_vars_w, used to store key-value pairs of file format-Pandas series of measurements. - Define a
forloop to iterate over the number of trialsn. - Define a writing method for each file format.
- Set a
starttimer variable before writing execution using thetime.time()method. This will be our initial time marker. - Set an
endtimer variable after writing execution using thetime.time()method. This will be our final time marker. - Calculate the
exec_timefor each loop by calculating the difference betweenendandstartvariables. - Append the
exec_timeto our correspondingpandas.core.series.Seriesobject. - Upon loop completion, assign a key-value pair to our
measured_vars_wdictionary corresponding to the file format and measured execution times. - Remove all generated files except one using the
os.removemethod. - Repeat for all remaining file formats.
We can translate our pseudocode into code:
Code
def writingPerformance(n, path, df):
'''
Parameters
----------
n : int
Number of trials.
path : str
Path for result writing.
df : pandas.DataFrame
DataFrame Object containing data set.
Returns
-------
measured_vars_w : dict
Execution time for each file format, with n number of trials.
'''
# Declare pd.Series() for storing the measured variables for each file format
wtime_csv = pd.Series([], dtype='float64')
wtime_txt = pd.Series([], dtype='float64')
wtime_feather = pd.Series([], dtype='float64')
wtime_parquet_NP = pd.Series([], dtype='float64')
wtime_parquet_SP = pd.Series([], dtype='float64')
wtime_parquet_MP = pd.Series([], dtype='float64')
wtime_avro = pd.Series([], dtype='float64')
wtime_pickle = pd.Series([], dtype='float64')
# Declare a dictionary for storing all series
measured_vars_w = {}
# -------------------------------
# 1. CSV
# -------------------------------
for trial in range(n):
# Start trial
print(f'CSV trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to file
df.to_csv(path + 'CSV' + '_' + str(trial) + '.csv')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_csv = pd.concat([pd.Series([exec_time]), wtime_csv])
# Define series title
wtime_csv.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['01_CSV'] = wtime_csv.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
os.remove(path + 'CSV' + '_' + str(trial) + '.csv')
# -------------------------------
# 2. TXT
# -------------------------------
for trial in range(n):
# Start trial
print(f'TXT trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to file
df.to_csv(path + 'TXT' + '_' + str(trial) + '.txt', sep = '\t')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_txt = pd.concat([pd.Series([exec_time]), wtime_txt])
# Define series title
wtime_txt.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['02_TXT'] = wtime_txt.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
os.remove(path + 'TXT' + '_' + str(trial) + '.txt')
# -------------------------------
# 3. Feather
# -------------------------------
for trial in range(n):
# Start trial
print(f'Feather trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to file
df.to_feather(path + 'Feather' + '_' + str(trial) + '.feather')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_feather = pd.concat([pd.Series([exec_time]), wtime_feather])
# Define series title
wtime_feather.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['03_Feather'] = wtime_feather.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
os.remove(path + 'Feather' + '_' + str(trial) + '.feather')
# -------------------------------
# 4. Parquet non-partitioned
# -------------------------------
for trial in range(n):
# Start trial
print(f'Parquet NP trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to file
df.to_parquet(path + 'Parquet_NP' + '_' + str(trial) + '.parquet')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_parquet_NP = pd.concat([pd.Series([exec_time]), wtime_parquet_NP])
# Define series title
wtime_parquet_NP.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['04_Parquet_NP'] = wtime_parquet_NP.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
os.remove(path + 'Parquet_NP' + '_' + str(trial) + '.parquet')
# -------------------------------
# 5. Parquet single-partitioned
# -------------------------------
for trial in range(n):
# Start trial
print(f'Parquet SP trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to file
df.to_parquet(path + 'Parquet_SP' + '_' + str(trial) + '.parquet', partition_cols = ['region'])
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_parquet_SP = pd.concat([pd.Series([exec_time]), wtime_parquet_SP])
# Define series title
wtime_parquet_SP.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['05_Parquet_SP'] = wtime_parquet_SP.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
shutil.rmtree(path + 'Parquet_SP' + '_' + str(trial) + '.parquet')
# -------------------------------
# 6. Parquet multi-partitioned
# -------------------------------
for trial in range(n):
# Start trial
print(f'Parquet MP trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to file
df.to_parquet(path + 'Parquet_MP' + '_' + str(trial) + '.parquet', partition_cols = ['region', 'trend'])
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_parquet_MP = pd.concat([pd.Series([exec_time]), wtime_parquet_MP])
# Define series title
wtime_parquet_MP.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['06_Parquet_MP'] = wtime_parquet_MP.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
shutil.rmtree(path + 'Parquet_MP' + '_' + str(trial) + '.parquet')
# -------------------------------
# 7. Avro
# -------------------------------
# Define the schema
schema = {
'type': 'record',
'name': 'performance_comp',
'namespace': 'performance_comp',
'doc': 'This schema consists of 2 int types, 1 datetime type and 6 string types',
'fields': [
{'name': 'title', 'type': 'string'},
{'name': 'rank', 'type': 'int'},
{'name': 'date', 'type': 'long'},
{'name': 'artist', 'type': 'string'},
{'name': 'url', 'type': 'string'},
{'name': 'region', 'type': 'string'},
{'name': 'chart', 'type': 'string'},
{'name': 'trend', 'type': 'string'},
{'name': 'streams', 'type': 'int'}
]
}
# Parse the schema
parsed_schema = parse_schema(schema)
# Convert pd.DataFrame to records (list of dictionaries)
records = df.to_dict('records')
for trial in range(n):
# Start trial
print(f'Avro trial {trial} of {n} started...')
# Start timer
start = time.time()
# Write to Avro file
with open(path + 'Avro' + '_' + str(trial) + '.avro', 'wb') as file:
writer(file, parsed_schema, records)
file.close()
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_avro = pd.concat([pd.Series([exec_time]), wtime_avro])
# Define series title
wtime_avro.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['07_Avro'] = wtime_avro.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
os.remove(path + 'Avro' + '_' + str(trial) + '.avro')
# -------------------------------
# 8. Pickle open file
# -------------------------------
for trial in range(n):
# Start trial
print(f'Pickle trial {trial} of {n} started...')
# Convert pd.DataFrame to records (list of dictionaries)
records = df.to_dict('records')
# Start timer
start = time.time()
file = open(path + 'Pickle' + '_' + str(trial) + '.pickle', 'wb')
# Write open file to disk
pickle.dump(records, file)
file.close()
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
wtime_pickle = pd.concat([pd.Series([exec_time]), wtime_pickle])
# Define series title
wtime_pickle.name = 'Writing Time [s]'
# Add measurements to dictionary
measured_vars_w['08_Pickle'] = wtime_pickle.reset_index(drop = True)
# Remove all files from dir except one
for trial in range(n - 1):
os.remove(path + 'Pickle' + '_' + str(trial) + '.pickle')
return measured_vars_w3. Reading performance tests
Similar to the writing performance tests section, we will start by defining our readingPerformance() function, which will accept the following parameters :
n:int- Number of trials.
path:str- Path for writing results.
df:pandas.DataFrame- DataFrame Object containing data set.
Upon calling, it will return the following:
measured_vars_r:dict- Execution time for each file format, with
nnumber of trials.
- Execution time for each file format, with
Once we have the expected inputs and outputs, the idea is to perform the following:
- Declare a
pandas.core.series.Seriesfor each file format, where we will store writing times. This would result in 8 objects in total. - Declare an empty dictionary
measured_vars_w, used to store key-value pairs of file format-Pandas series of measurements. - Define a
forloop to iterate over the number of trialsn. - Define a reading method for each file format.
- Set a
starttimer variable before writing execution using thetime.time()method. This will be our initial time marker. - Set an
endtimer variable after writing execution using thetime.time()method. This will be our final time marker. - Calculate the
exec_timefor each loop by calculating the difference betweenendandstartvariables. - Append the
exec_timeto our correspondingpandas.core.series.Seriesobject. - Delete the read object from memory.
- Upon loop completion, assign a key-value pair to our
measured_vars_wdictionary corresponding to the file format and measured execution times. - Repeat for all remaining file formats.
We can translate our pseudocode into code:
Code
def readingPerformance(n, path, df):
'''
Parameters
----------
n : int
Number of trials.
path : str
Path for result writing.
df : pandas.DataFrame
DataFrame Objectn containing data set.
Returns
-------
measured_vars_r : dict
Execution time for each file format, with n number of trials.
'''
# Declare pd.Series() for storing the measured variables for each file format
rtime_csv = pd.Series([], dtype='float64')
rtime_txt = pd.Series([], dtype='float64')
rtime_feather = pd.Series([], dtype='float64')
rtime_parquet_NP = pd.Series([], dtype='float64')
rtime_parquet_SP = pd.Series([], dtype='float64')
rtime_parquet_MP = pd.Series([], dtype='float64')
rtime_avro = pd.Series([], dtype='float64')
rtime_pickle = pd.Series([], dtype='float64')
# Declare a dictionary for storing all series
measured_vars_r = {}
# -------------------------------
# 1. CSV
# -------------------------------
for trial in range(n):
# Start trial
print(f'CSV trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
df = pd.read_csv(path + 'CSV' + '_' + str(n-1) + '.csv')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_csv = pd.concat([pd.Series([exec_time]), rtime_csv])
# Delete file from memory
del df
# Define series title
rtime_csv.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['01_CSV'] = rtime_csv.reset_index(drop = True)
# -------------------------------
# 2. TXT
# -------------------------------
for trial in range(n):
# Start trial
print(f'TXT trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
df = pd.read_csv(path + 'TXT' + '_' + str(n-1) + '.txt', sep = '\t')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_txt = pd.concat([pd.Series([exec_time]), rtime_txt])
# Delete file from memory
del df
# Define series title
rtime_txt.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['02_TXT'] = rtime_txt.reset_index(drop = True)
# -------------------------------
# 3. Feather
# -------------------------------
for trial in range(n):
# Start trial
print(f'Feather trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
df = pd.read_feather(path + 'Feather' + '_' + str(n-1) + '.feather')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_feather = pd.concat([pd.Series([exec_time]), rtime_feather])
# Delete file from memory
del df
# Define series title
rtime_feather.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['03_Feather'] = rtime_feather.reset_index(drop = True)
# -------------------------------
# 4. Parquet non-partitioned
# -------------------------------
for trial in range(n):
# Start trial
print(f'Parquet NP trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
df = pd.read_parquet(path + 'Parquet_NP' + '_' + str(n-1) + '.parquet')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_parquet_NP = pd.concat([pd.Series([exec_time]), rtime_parquet_NP])
# Delete file from memory
del df
# Define series title
rtime_parquet_NP.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['04_Parquet_NP'] = rtime_parquet_NP.reset_index(drop = True)
# -------------------------------
# 5. Parquet single-partitioned
# -------------------------------
for trial in range(n):
# Start trial
print(f'Parquet SP trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
df = pd.read_parquet(path + 'Parquet_SP' + '_' + str(n-1) + '.parquet')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_parquet_SP = pd.concat([pd.Series([exec_time]), rtime_parquet_SP])
# Delete file from memory
del df
# Define series title
rtime_parquet_SP.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['05_Parquet_SP'] = rtime_parquet_SP.reset_index(drop = True)
# -------------------------------
# 6. Parquet multi-partitioned
# -------------------------------
for trial in range(n):
# Start trial
print(f'Parquet MP trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
df = pd.read_parquet(path + 'Parquet_MP' + '_' + str(n-1) + '.parquet')
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_parquet_MP = pd.concat([pd.Series([exec_time]), rtime_parquet_MP])
# Delete file from memory
del df
# Define series title
rtime_parquet_MP.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['06_Parquet_MP'] = rtime_parquet_MP.reset_index(drop = True)
# -------------------------------
# 7. Avro
# -------------------------------
for trial in range(n):
# Start trial
print(f'Avro trial {trial} of {n} started...')
# Define list of dictionaries
lod = []
# Start timer
start = time.time()
# Read from Avro file
with open(path + 'Avro' + '_' + str(n-1) + '.avro', 'rb') as fo:
avro_reader = reader(fo)
for record in avro_reader:
lod.append(record)
# Close the BufferedReader object
fo.close()
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_avro = pd.concat([pd.Series([exec_time]), rtime_avro])
# Delete file from memory
del lod
# Define series title
rtime_avro.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['07_Avro'] = rtime_avro.reset_index(drop = True)
# -------------------------------
# 8. Pickle open file
# -------------------------------
for trial in range(n):
# Start trial
print(f'Pickle trial {trial} of {n} started...')
# Start timer
start = time.time()
# Read from file
with open(path + 'Pickle' + '_' + str(n-1) + '.pickle', 'rb') as file:
my_pickled_object = pickle.load(file)
# Close the BufferedReader object
file.close()
# End timer
end = time.time()
# Calculate execution time
exec_time = end - start
# Append time to series
rtime_pickle = pd.concat([pd.Series([exec_time]), rtime_pickle])
# Delete file from memory
del my_pickled_object
# Define series title
rtime_pickle.name = 'Reading Time [s]'
# Add measurements to dictionary
measured_vars_r['08_Pickle'] = rtime_pickle.reset_index(drop = True)
return measured_vars_r4. Analysis
Once we have both the writingPerformance() and readingPerformance() functions declared, we can define an analysis() function which will accept the following parameters:
n:int- Number of trials.
path:str- Path for writing results.
df:pandas.DataFrame- DataFrame Object containing data set.
Upon calling, it will return the following:
measured_vars_w:dict- Writing time of each file format with
nnumber of trials.
- Writing time of each file format with
measured_vars_r:dict- Reading time of each file format with
nnumber of trials.
- Reading time of each file format with
stat_dw:dict- Statistical description of measured writing times for all formats.
stat_dr:dict- Statistical description of measured reading times for all formats.
size_d:dict- Statistical description of measured file/folder sizes for all formats.
Once we have the expected inputs and outputs, the idea is to perform the following:
4.1 Writing performance analysis
- Define a list
testscontaining the names of each file format. We will use this object as iterable. - Call the
writingPerformance()function and assign it to ameasured_vars_wobject. - Define an empty dictionary
stat_dwfor saving the statistical descriptions for the writing test. - Iterate over the
testsobject, indexing themeasured_vars_wdictionary on each loop and calculating its statistical description. - Assign a key-value pair on each loop, consisting of the file format name as the key and the statistical description object as the value.
4.2 Reading performance analysis
- Call the
readingPerformance()function and assign it to ameasured_vars_robject. - Define an empty dictionary
stat_drfor saving the statistical descriptions for the reading test. - Iterate over the
testsobject, indexing themeasured_vars_rdictionary on each loop and calculating its statistical description. - Assign a key-value pair on each loop, consisting of the file format name as the key and the statistical description object as the value.
4.3 File size analysis
- Declare an empty dictionary
size_dfor storing the file size values. - For each file, calculate its size using the
os.path.getsizemethod. - For each folder (parquet partitions), recursively calculate its size using the
f.stat().st_sizemethod. - Convert file sizes:
- The
os.path.getsizemethod returns the file size in bytes, so we need to divide the result by 1,0242 to get our measurements inMBunits. - The
f.stat().st_sizemethod returns the folder size in bytes, so we need to divide the result by 1,0242 to get our measurements inMBunits.
- The
We can translate our pseudocode into code:
Code
def analysis(n, path, df):
'''
Parameters
----------
n : int
Number of trials.
path : str
Path for result writing.
df : pandas.DataFrame
DataFrame Objectn containing data set.
Returns
-------
measured_vars_w : dict
Writing time of each file format, with n number of trials.
measured_vars_r : dict
Reading time of each file format, with n number of trials.
stat_dw : dict
Statistical description of measured writing times for all formats.
stat_dr : dict
Statistical description of measured reading times for all formats.
size_d : dict
Statistical description of measured file/folder sizes for all formats.
'''
# Define all tests for including in statistical description
tests = ['01_CSV',
'02_TXT',
'03_Feather',
'04_Parquet_NP',
'05_Parquet_SP',
'06_Parquet_MP',
'07_Avro',
'08_Pickle']
# -------------------------------
# Writing Analysis
# -------------------------------
# Perform writing experiment and get measured_vars_w dictionary
measured_vars_w = writingPerformance(n, path, df)
# Define statistical results dict
stat_dw = {}
# Statistical Description for Writing
# Extract each set of values, describe them and save them in a new dict
for i in tests:
stat_v = measured_vars_w[i].describe()
stat_dw[i] = stat_v
# -------------------------------
# Reading Analysis
# -------------------------------
# Perform reading experiment and get measured_vars_r dictionary
measured_vars_r = readingPerformance(n, path, df)
# Define statistical results dict
stat_dr = {}
# Statistical Description for Reading
# Extract each set of values, describe them and save them in a new dict
for i in tests:
stat_v = measured_vars_r[i].describe()
stat_dr[i] = stat_v
# -------------------------------
# File Size Analysis
# -------------------------------
# Define size results dict
size_d = {}
# Calculate file & dir sizes
size_d['01_CSV'] = os.path.getsize(path + 'CSV' + '_' + str(n-1) + '.csv') / (1024**2)
size_d['02_TXT'] = os.path.getsize(path + 'TXT' + '_' + str(n-1) + '.txt') / (1024**2)
size_d['03_Feather'] = os.path.getsize(path + 'Feather' + '_' + str(n-1) + '.feather') / (1024**2)
size_d['04_Parquet_NP'] = os.path.getsize(path + 'Parquet_NP' + '_' + str(n-1) + '.parquet') / (1024**2)
path_Parquet_SP = Path(path + 'Parquet_SP_19.parquet')
size_d['05_Parquet_SP'] = sum(f.stat().st_size for f in path_Parquet_SP.glob('**/*') if f.is_file()) / (1024**2)
path_Parquet_MP = Path(path + 'Parquet_MP_19.parquet')
size_d['06_Parquet_MP'] = sum(f.stat().st_size for f in path_Parquet_MP.glob('**/*') if f.is_file()) / (1024**2)
size_d['07_Avro'] = os.path.getsize(path + 'Avro' + '_' + str(n-1) + '.avro') / (1024**2)
size_d['08_Pickle'] = os.path.getsize(path + 'Pickle' + '_' + str(n-1) + '.pickle') / (1024**2)
# Return a dictionary including the actual measured time values of all methods
# Return a dictionary including the statistical description of all methods
return measured_vars_w, measured_vars_r, stat_dw, stat_dr, size_dIf we closely examine lines 1049 through 1059, we see that we need to declare different methods for calculating file sizes & folder sizes.
We can then call our analysis() function and assign the outputs to 5 different objects:
Code
# Call analysis function
measured_vars_w, measured_vars_r, stat_dw, stat_dr, size_d = analysis(n, path, df)
# Print the type and shape of each object
print(type(measured_vars_w))
print(len(measured_vars_w))
print(type(measured_vars_r))
print(len(measured_vars_r))
print(type(stat_dw))
print(len(stat_dw))
print(type(stat_dr))
print(len(stat_dr))
print(type(size_d))
print(len(size_d))Output
<class 'dict'>
8
<class 'dict'>
8
<class 'dict'>
8
<class 'dict'>
8
<class 'dict'>
8The shapes of our objects match the output requirements, as we have eight tested file formats.
Performance results
We now have everything we need to make sense of the data we just generated.
In this example, the quantity of data is manageable since we only have eight file formats and 20 measurements per format. Regardless, it is always a good practice to visualize the results in a plot as a first step.
1. Plotting the results
We’ll proceed with some visualization techniques appropriate for the results we have.
1.1 Bar chart for file sizes
A bar chart is a simple way to visualize data quickly. We can generate a bar chart using the matplotlib.pyplot module:
Code
# Create figure
plt.figure('File Sizes Bar Chart')
# Plot the file sizes
plt.bar(size_d.keys(), size_d.values(), color = color_main)
# Enable grid
plt.grid(True, zorder=0)
# Set xlabel and ylabel
plt.xlabel('File Format', fontsize=label_font_size, labelpad=text_padding)
plt.ylabel('File Size [GB]', fontsize=label_font_size, labelpad=text_padding)
# Remove bottom and top separators
sns.despine(bottom=True)
# Add plot title
plt.title('File/Folder Sizes in Gigabytes', fontsize=title_font_size, pad=text_padding)
# Optional: Save the figure as a png image
plt.savefig('performance_results/' + 'file_sizes_bar_chart_tp.png', format = 'png', dpi = 300, transparent = True)
plt.savefig('performance_results/' + 'file_sizes_bar_chart_bg.png', format = 'png', dpi = 300, transparent = False)
# Close the figure
plt.close()If we save the figure directly and then close it, the image will be written in the path we specify and will not display on our IDE.
If we examine line 1181, we used an additional library called seaborn to include the additional parameter sns.despine(). This module is handy when dealing with statistical analysis visualizations. However, we will not use it in this particular section and will limit ourselves to using the matplotlib.pyplot module.
Output
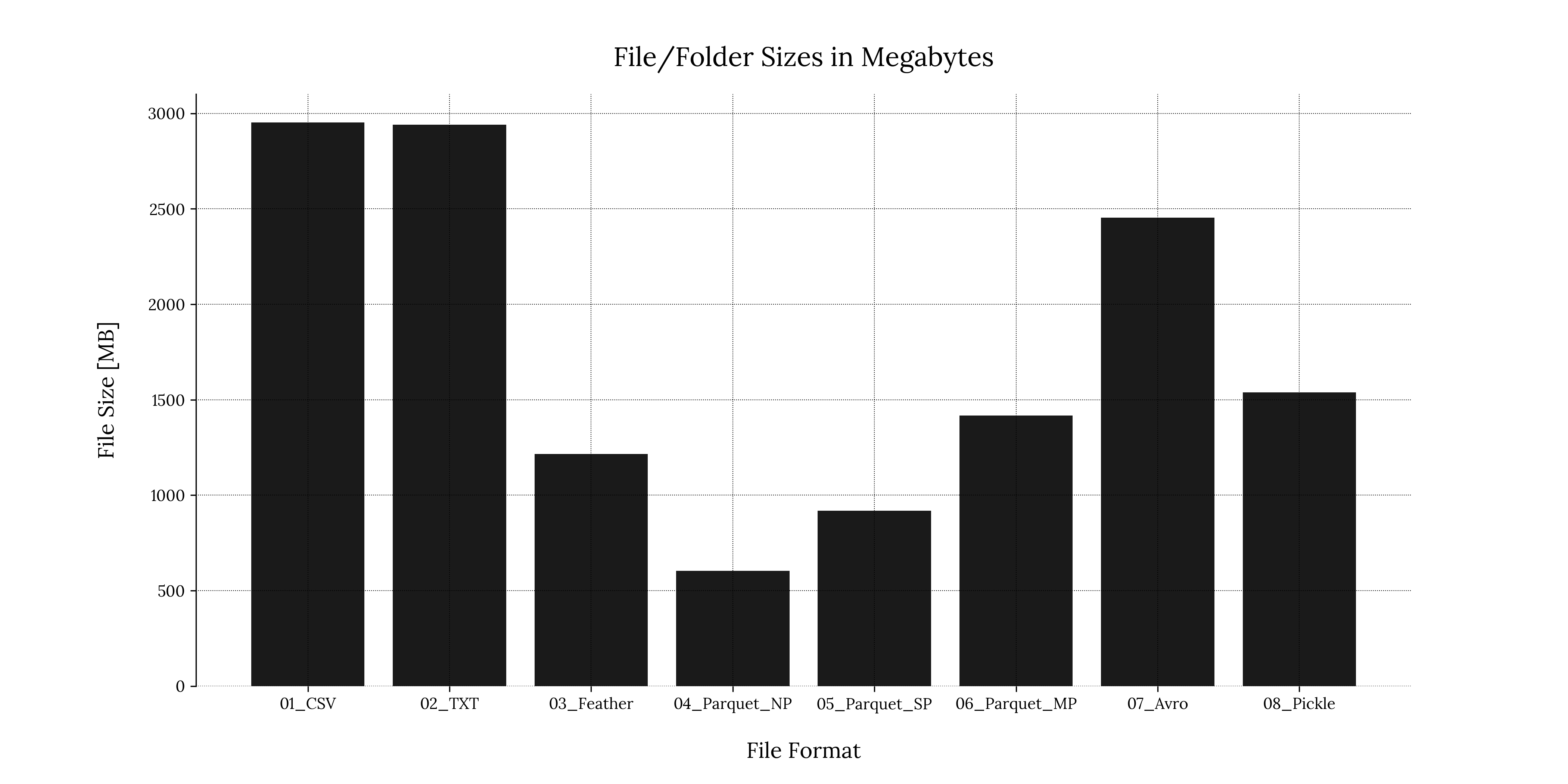
MB for each file format1.2 Boxplot for writing times
A boxplot is a visualization method widely used in Data Science & statistical analysis. Its purpose is to describe the distribution of experimental measurements, including useful visual information about the set.
A detailed discussion of the boxplot components is out of the scope of this article. That will be covered in another blog post.
We can generate a boxplot using the matplotlib.pyplot module:
Code
# Create figure
plt.figure('Writing Times Boxplot')
# Plot the writing times
plt.boxplot(measured_vars_w.values(),
labels = measured_vars_w.keys(),
showmeans=True)
# Enable grid
plt.grid(True, zorder=0)
# Set xlabel and ylabel
plt.xlabel("File Format", fontsize=label_font_size, labelpad=text_padding)
plt.ylabel("Writing Time [s]", fontsize=label_font_size, labelpad=text_padding)
# Remove bottom and top separators
sns.despine(bottom=True)
# Add plot title
plt.title('Writing Time in Seconds', fontsize=title_font_size, pad=text_padding)
# Optional: Save the figure as a png image
plt.savefig('performance_results/' + 'writing_time_scattered_boxplots_tp.png', format = 'png', dpi = 300, transparent = True)
plt.savefig('performance_results/' + 'writing_time_scattered_boxplots_bg.png', format = 'png', dpi = 300, transparent = False)
# Close the figure
plt.close()Output
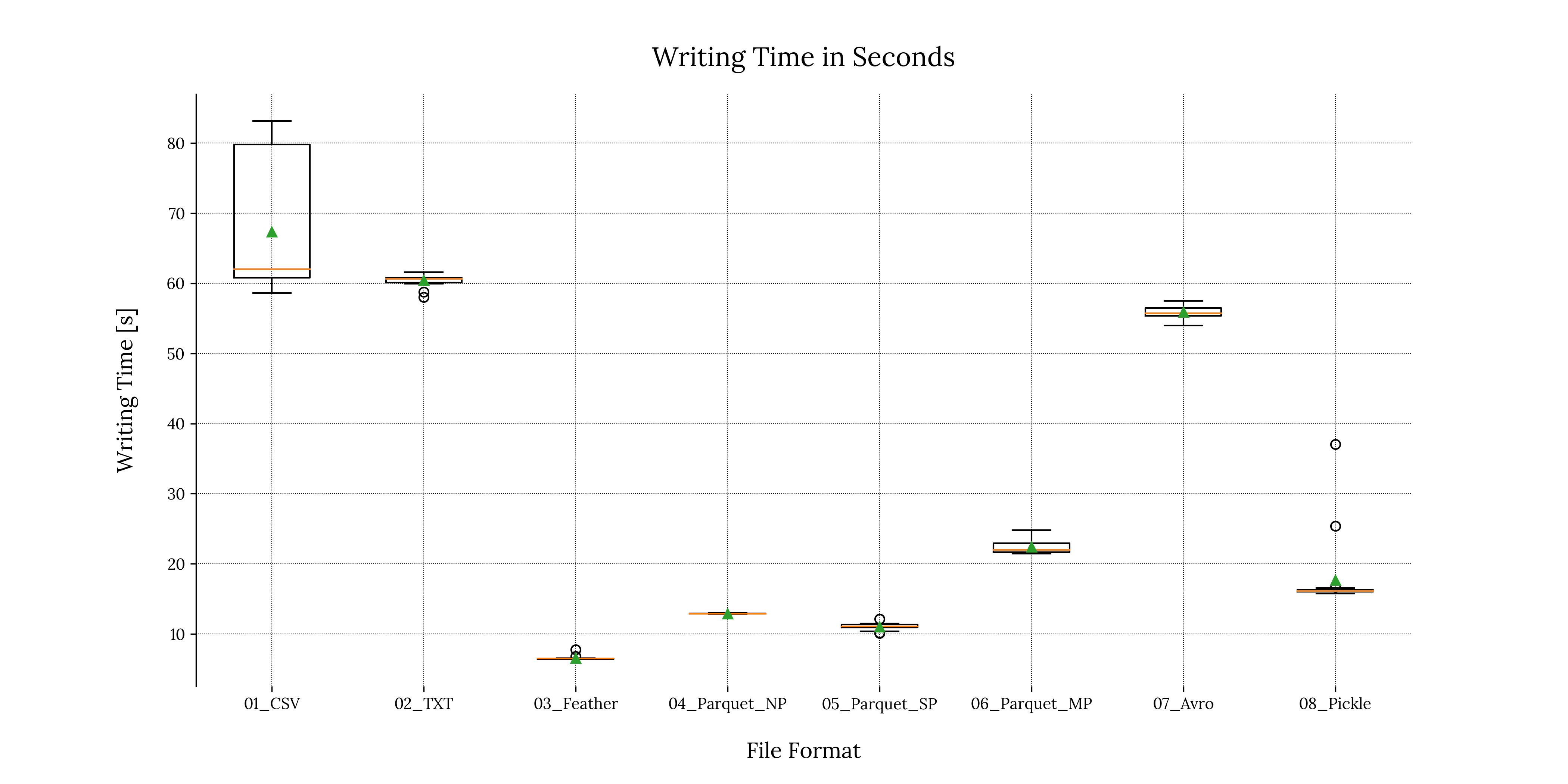
1.3 Boxplot for reading times
We can perform a similar treatment to our reading time results:
Code
# Create figure
plt.figure('Reading Times Boxplot')
# Plot the writing times
plt.boxplot(measured_vars_r.values(),
labels = measured_vars_r.keys(),
showmeans=True)
# Enable grid
plt.grid(True, zorder=0)
# Set xlabel and ylabel
plt.xlabel("File Format", fontsize=label_font_size, labelpad=text_padding)
plt.ylabel("Reading Time [s]", fontsize=label_font_size, labelpad=text_padding)
# Remove bottom and top separators
sns.despine(bottom=True)
# Add plot title
plt.title('Reading Time in Seconds', fontsize=title_font_size, pad=text_padding)
# Optional: Save the figure as a png image
plt.savefig('performance_results/' + 'reading_time_scattered_boxplots_tp.png', format = 'png', dpi = 300, transparent = True)
plt.savefig('performance_results/' + 'reading_time_scattered_boxplots_bg.png', format = 'png', dpi = 300, transparent = False)
# Close the figure
plt.close()Output
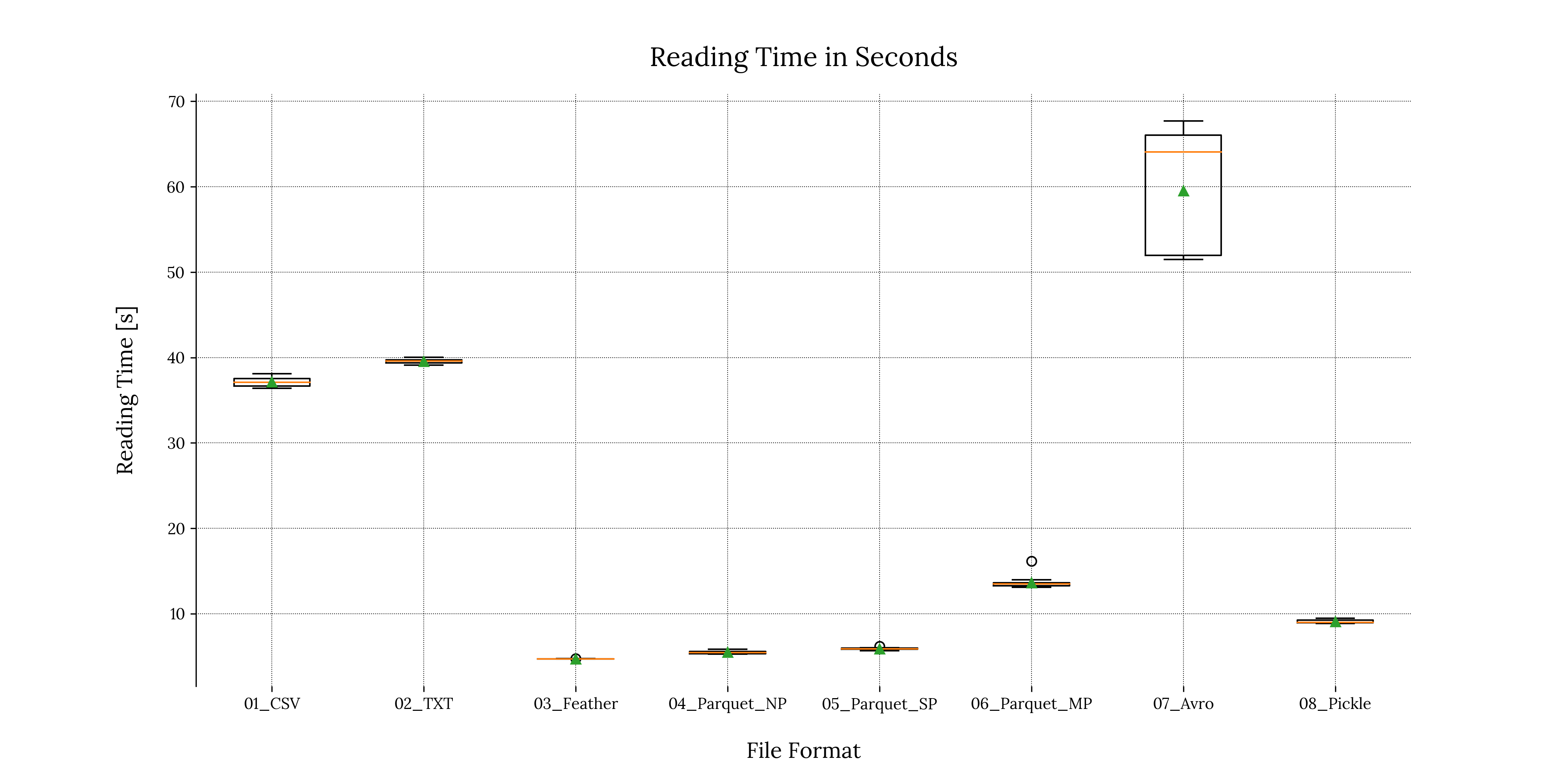
2. Exporting the results in a tabular format
We can also write our results in an Excel file. This is a valuable technique whenever we want to share or store information that took a fair amount of time to generate (imagine explaining to our boss why we had to re-run a 2-hour performance test just to get the results back).
An Excel file is also a very friendly tabular format that everyone understands. It can be used to make further analyses such as pivoting or calculating statistical measures (![]() ,
, ![]() ,
, ![]() ,
, ![]() , among others).
, among others).
For this part, we will be writing four .xlsx files, two for writing results and two for reading results. Each file will have eight tabs, each consisting of the file format and the writing and reading times in seconds, respectively:
Code
# Define function to export results to Excel file
def results_to_excel(dseries_dict, path):
"""Write dictionary of dataframes to separate sheets, within
1 file."""
writer = pd.ExcelWriter(path, engine='openpyxl')
for tab_name, dseries in dseries_dict.items():
dseries.to_excel(writer, sheet_name=tab_name)
writer.close()
# Define file for writing results
path_w = 'performance_results/' + 'measured_vars_w.xlsx'
path_dw = 'performance_results/' + 'stat_dw.xlsx'
# Call function on writing results
results_to_excel(measured_vars_w, path_w)
results_to_excel(stat_dw, path_dw)
# Define file for reading results
path_r = 'performance_results/' + 'measured_vars_r.xlsx'
path_dr = 'performance_results/' + 'stat_dr.xlsx'
# Call function on reading results
results_to_excel(measured_vars_r, path_r)
results_to_excel(stat_dr, path_dr)Side-by-side comparison
1. Consolidated results
| Format | Size [MB] | Avg. Writing Time [s] | Writing Method | Avg. Reading Time [s] | Reading Method |
.csv | 2,954 | 67.4 | df.to_csv() | 37.1 | pd.read_csv() |
.txt | 2,941 | 60.4 | df.to_csv() | 39.6 | pd.read_csv() |
.feather | 1,216 | 6.6 | df.to_feather() | 4.7 | pd.read_feather() |
.parquet | 604 | 12.9 | df.to_parquet() (NP) | 5.5 | pd.read_parquet() |
.parquet | 919 | 11.0 | df.to_parquet() (SP) | 5.9 | pd.read_parquet() |
.parquet | 1,417 | 22.4 | df.to_parquet() (MP) | 13.6 | pd.read_parquet() |
.avro | 2,455 | 55.9 | fastavro writer() | 59.5 | fastavro reader() |
.pickle | 1,539 | 17.6 | pickle.dump() | 9.1 | pickle.load() |
Note: Keep in mind that these values vary across systems. CPU processing power, RAM capacity, and other variables directly affect reading & writing times. Please refer to the Appendix section for the full list of machine specifications used in this experiment.
2. Interpretation
We can see that the .csv & .txt file formats had the largest file sizes, while the non-partitioned .parquet had the smallest file size. .parquet folder sizes increased almost linearly as we increased the number of partitions, which makes sense if we remember that a partitioned file creates a directory hierarchy beneath the main directory.
For writing times, we can see that most of our measures were more or less consistent except with the .pickle file format. This is denoted by the two outliers visible in Figure 1.2 and can be due to processes starting in the background, thus reducing resources and introducing noise in the measurements. We can mitigate or at least reduce this phenomenon by tightening our experimental conditions.
Writing .feather files consistently consumed the least amount of time, followed by the single partitioned .parquet file and non-partitioned .parquet file consecutively. We can also clearly see that the .csv and .txt file formats were the worst performing, followed by the .avro format.
For reading times, the story is slightly different: Reading from the .avro file consumed the most amount of time (25 seconds more than the second worst case), along with the highest standard deviation value of all file formats (![]() , meaning 7 seconds of deviation from the mean). This is due to the fact that the
, meaning 7 seconds of deviation from the mean). This is due to the fact that the fastavro library provides an iterator reading object, meaning the script had to iterate over each row and append it to an object (in our case, a list). This additional step adds an extra layer of computational complexity, therefore increasing reading times.
In contrast, the .feather file format took the least amount of time to read, closely followed by the non-partitioned & single-partitioned .parquet files.
Use cases
From the results obtained in the previous section, we can see that the .feather file format offers consistently fast writing & reading speeds when compared to the other file formats. Also, the file size is considerably smaller. However, as we previously mentioned, this format is not recommended for long-term storage due to its binary form instability.
A great alternative to handling big data would be the stabler .parquet file format: Both non-partitioned and single-partitioned forms presented low writing & reading speeds and relatively small file sizes compared to the other formats.
Even though the non-partitioned approach was the overall highest performing, it is not always the best option: when dealing with large data sets that contain multiple aggregation levels, partitioning lets us divide the aggregation levels into subfolders, making the information accessible by blocks, meaning we would not require to read the entire file to extract a single column field group. This is especially important with big data since Python reads data and stores it in memory, so a single-partitioned 1TB .parquet file would be practically impossible to read (parsing by blocks could do the trick, but then, this is what .parquet partitioning achieves more elegantly, right?).
Another high-performing case was the .pickle file format, but we did not mention it as a first or even second option for two extremely relevant reasons:
- It’s platform specific.
- Malicious code can be easily injected, creating potential vulnerabilities in our environment.
These two reasons alone make .pickle files more of a niche solution for interchanging Python objects and most definitely not apt for production environments.
Lastly, the two worst performing file formats, .csv and .txt, which ironically are the most popular, present all of the advantages that we mentioned earlier, but have substantial limitations if we want to ensure a proper and secure data writing & loading environment; the single fact that these two file formats don’t preserve data types is a good-enough reason to evaluate other alternatives.
Conclusions
In summary, not all “big data” file formats are tailored for handling big data. This is counterintuitive, nonetheless true. Each was created with a specific purpose and is better or worse at some applications than others.
Serialized formats, for example, present several advantages over non-serialized formats. Still, the inverse also happens: serialized formats may lose stability across versions and usually consume more processing resources when serializing/deserializing objects.
Before implementing a format, especially in a production environment, it’s essential to make a detailed assessment of which formats will be used and how the encoding will be handled. Also, when working with other collaborators, setting a strict standard for data formatting & handling is of vital importance and, more often than not, overlooked in favor of implementing the “easier way”.
Appendix
1. Experimental conditions
Below we can find a list of the parameters that were used to obtain these results:
- Python Build: Python 3.11.1 (tags/v3.11.1:a7a450f, Dec 6 2022, 19:58:39) MSC v.1934 64 bit (AMD64) on win32
- Operating System: Windows 11 Home, Version 22H2
- Processor: 12th Gen Intel Core i9-12900H, 2.50 GHz
- RAM: 64 GB DDR5

